ResBoxes
Residue
Boxes - A single letter code is shown, representing
the amino acids of the protein sequence which structure is inspected.
The amino acid Boxes are color coded according to either STING_Paint
code (1) or according to William
Taylor (2) code.
Placing the cursor above
this element: pop-up area will show the position (sequence number
and amino acid three letter code) for selected amino acid.
Left mouse click: no action
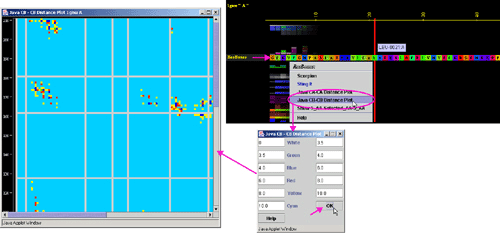
Right mouse
click: on any of the "ResBox" will generate following menu
and actions:
Figure below is showing specific position for the row with the "ResBox"
data in JPD presentation.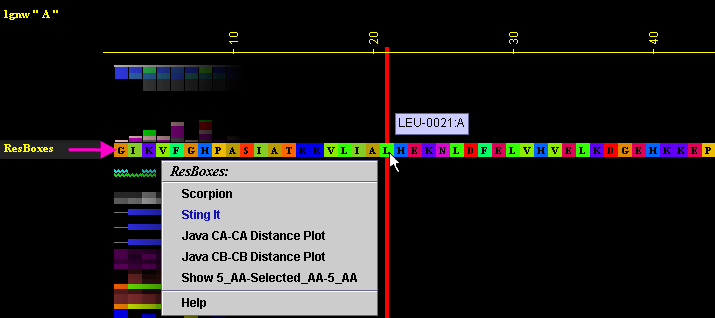
A) "Scorpion"
will call Scorpion component of BLUE STAR STING (see
HELP on Scorpion):
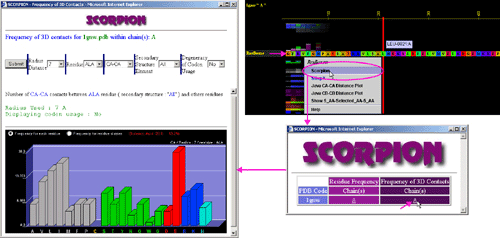
B) STING it - will induce
CPK rendering for selected amino acid.
Rendering
and color can be chosen among number of options
in the icons menu of JPD:
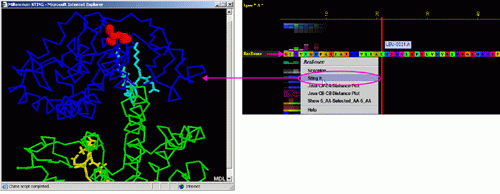
C) "Java Ca-Ca
Distance Plot" will call equally named component of BLUE STAR STING (see
HELP on Java Ca-Ca Distance
Plot ):
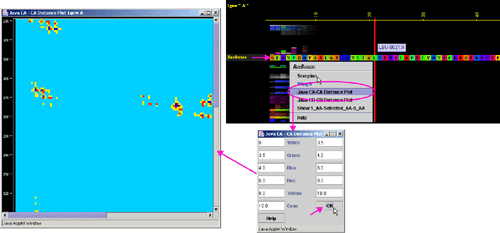
D) "Java Cb-Cb
Distance Plot" will call equally named component of BLUE STAR STING (see
HELP on Java Cb-Cb Distance
Plot ):
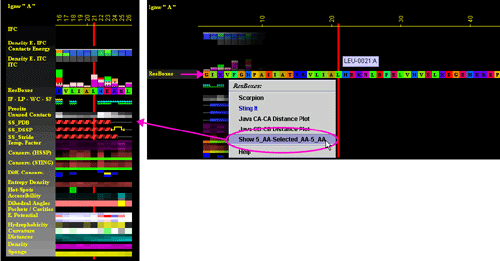
E) "Show 5_AA-Selected_AA-5_AA"
will show a portion of the JPD area with central amino acid and 5 amino
acids from each side:
For residues where the experimental technique (x-ray crystallography)
and its electron density does not permit amino acid identification, these
positions are presented as gaps or undetermined (see 1cho.pdb):
