Interface
Residues/Ligand Pocket residues/
Water Contacting residues/ Surface residues
(IF - LP - WC - SF)
InterFace area - Residues placed at the interface between two protein chains. Those residues are assigned after the calculation of the accessible surface area (using SurfV (10) program) of the protein chain in isolation and upon formation of the complex with the other(s) chain(s) present in the PDB file. If those two values are different, then selected amino acids suffered decrease in ASA and consequently belongs to the INTERFACE. The RED wiggly line representing IF residues changes the color to Blue when IF residues have just BARELY different values of ASA in isolation and in complex (see for such example in 15c8.pdb_D_170).
Placing the cursor above this element:pop-up area will show the position
(sequence number and amino acid three letter code) for selected amino
acid.
Left mouse click: no action
Right mouse
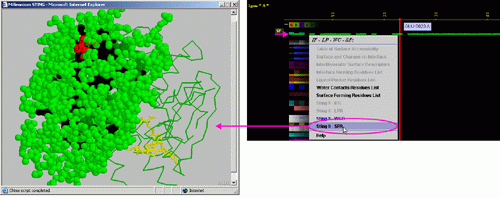
click: on any of the "IF" will generate following menu and
actions:
Figure below is showing
specific position for the row with the "IF" data in JPD presentation.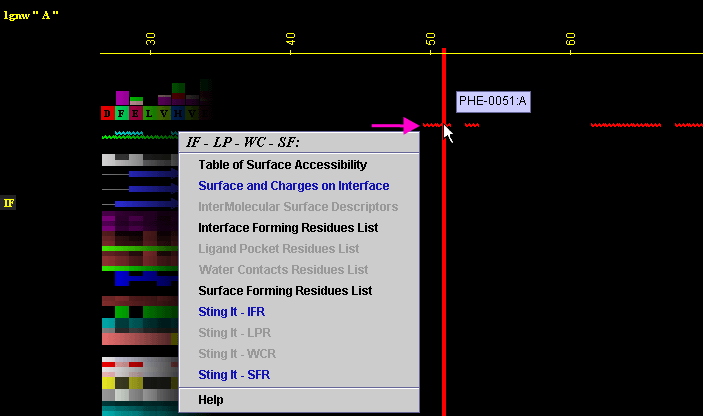
A) "Table of surface Accessibility " will call FORMIGA component
of BLUE STAR STING and by running the software SurfV, produce following table
(see
HELP on FORMIGA):
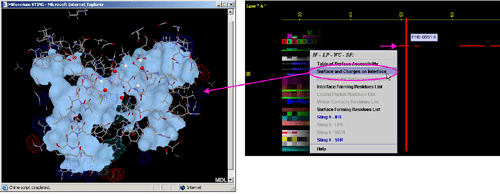
B) "Surface and Charges on Interface" will call specific Chime
script and produce following 3D output (see HELP on Chime Script):
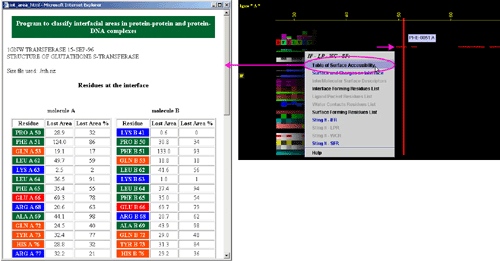
C) "Interface Forming Residue List" will call FORMIGA component
of BLUE STAR STING and by running the software SurfV, produce following table
(see HELP on
FORMIGA)(only amino acids with a difference in ASA value for
chain in isolation and chain in complex with the other(s) chain(s) present
in this particular PDB file, are presented):
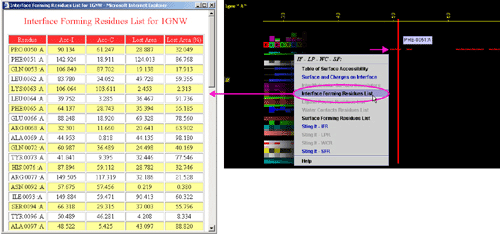
D) "Surface Forming Residue List" will call FORMIGA component
of BLUE STAR STING and by running the software SurfV, produce following table
(see
HELP on FORMIGA) (only amino acids with non zero ASA
are shown):
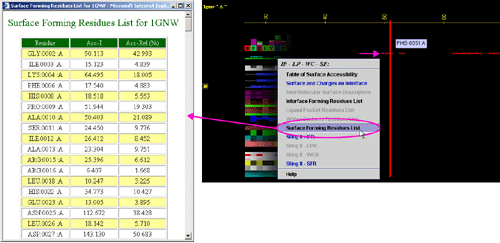
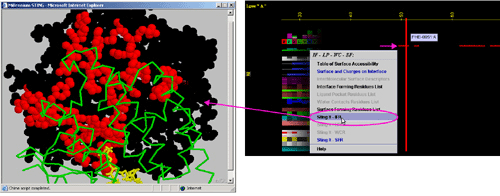
E) "STING it IFR " will call
a specific Chime script and produce following 3D output - showing
in red CPK only IFR residues:
F) "STING it SFR " will call
a specific Chime script and produce following 3D output - showing
in red CPK only SuRFace residues: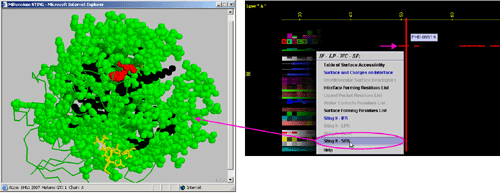
LP
(Ligand Pocket forming residues): Those are the amino acids which have
an identified atom at the distance not larger than the 4A from a ligand.
Placing the cursor above
this element:pop-up area will show the position (sequence number and
amino acid three letter code) for selected amino acid.
Left mouse click: no action
Right mouse
click: on any of the "LP" will generate following menu and
actions:
Figure below is showing
specific position for the row with the "LP" data in JPD presentation.
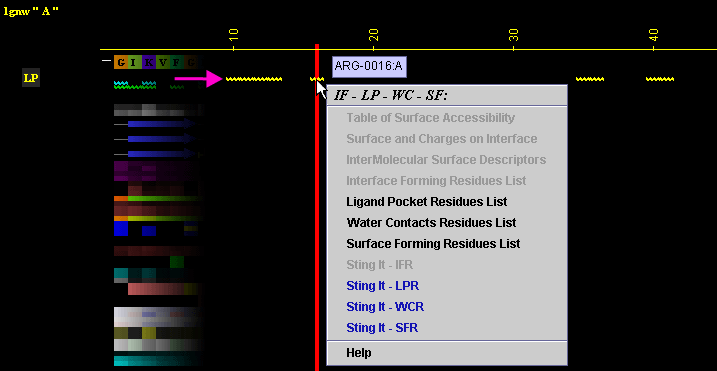
A) "Ligand Pocket
Residue List" will produce following table (only amino acids with
an atom identified at the distance not larger than 4A from the ligand
are shown):
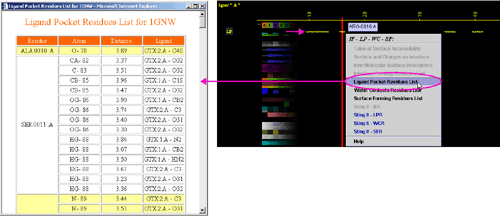
B) "Water Contacting Residues List" will produce following table
(only amino acids with an atom identified at the distance not larger than
4A from the co-crystalized water are shown):
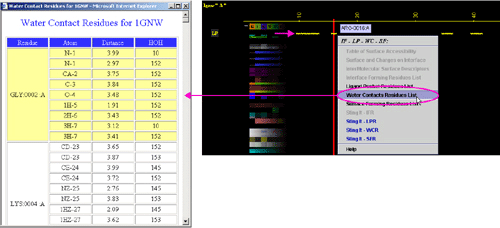
C) "Surface
Forming Residue List" will call FORMIGA component of BLUE STAR STING and
by running the software SurfV, produce following table (see
HELP on FORMIGA) (only amino acids with non zero ASA
are shown):
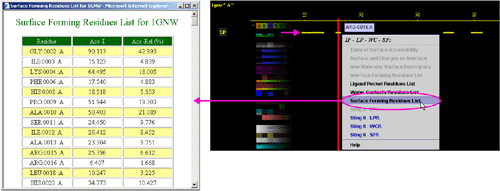
D) "STING it LPR
" will call a specific
Chime script and produce following 3D output - showing in yellow
CPK only amino
acids with an atom identified at the distance not larger than 4A from
the ligand :
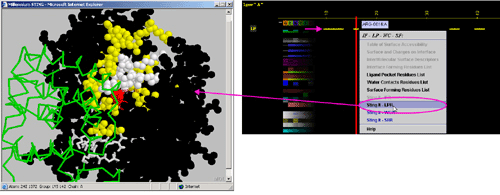
E) "STING it WCR " will call
a specific Chime script and produce following 3D output - showing
in cyan CPK only amino acids
with an atom identified at the distance not larger than 4A from the co-crystalized
water:
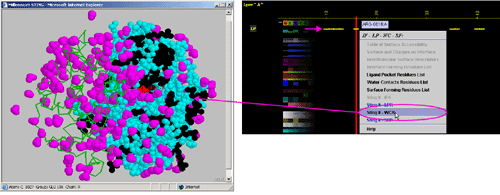
F) "STING it SFR " will call
a specific Chime script and produce following 3D output - showing
in red CPK only SuRFace residues:
WC
(Water Pocket forming residues): Those
are the amino acids which have an identified atom at the distance not
larger than the 4A from a water molecule (co-crystalized water molecules
present in the protein crystal).
Placing the cursor above
this element:pop-up area will show the position (sequence number and
amino acid three letter code) for selected amino acid.
Left mouse click: no action
Right mouse
click: on any of the "WC" will generate following menu and
actions:
Figure below is showing specific position for the row with the "WC"
data in JPD presentation.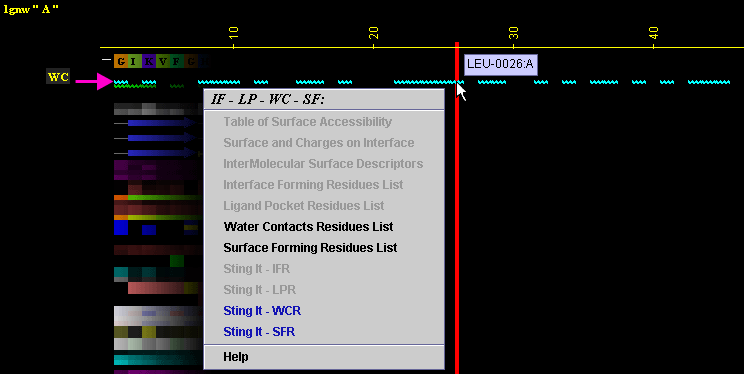
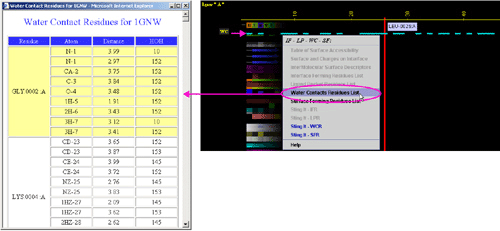
A) "Water Contacting
Residues List" will produce following table (only amino acids with
an atom identified at the distance not larger than 4A from the co-crystalized
water are shown):
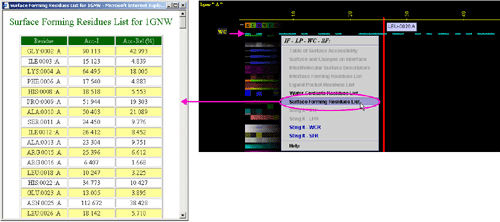
B) "Surface Forming Residue List" will call FORMIGA component
of BLUE STAR STING and by running the software SurfV, produce following table
(see
HELP on FORMIGA) (only amino acids with non zero ASA
are shown):
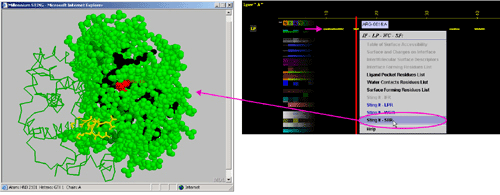
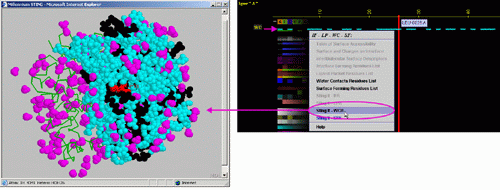
C) "STING it WCR " will call
a specific Chime script and produce following 3D output - showing
in cyan CPK only amino acids
with an atom identified at the distance not larger than 4A from the co-crystalized
water:
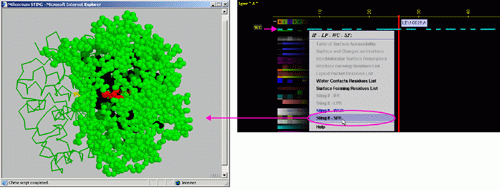
D) "STING it SFR " will call
a specific Chime script and produce following 3D output - showing
in red CPK only SuRFace residues:
SF
( surface forming residues): Those
are the amino acids which have accessible surface area >0. The amino
acids with accessible surface area numerical value from 0.01 to 3 A2
are underlined with the darker green color wiggly line.
Residues are selected into this subset if their ASA is >3 A.
The ASA value is calculated using the
SurfV (10)
program.
Placing the
cursor above this element:pop-up area will show the position (sequence
number and amino acid three letter code) for selected amino acid.
Left mouse click: no action
Right mouse
click: on any of the "SF" will generate following menu and
actions:
Figure below is showing specific position for the row with the "SF"
data in JPD presentation.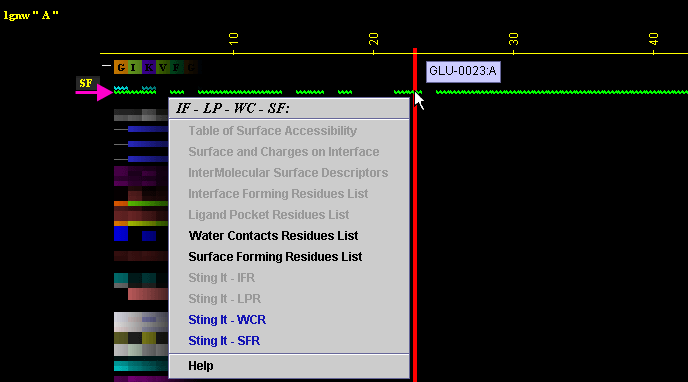
A) "Water Contacting Residues List" will produce following table
(only amino acids with an atom identified at the distance not larger than
4A from the co-crystalized water are shown):
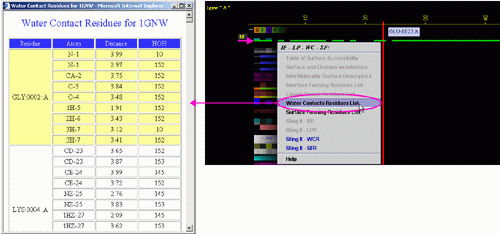
B) "Surface Forming Residue List" will call FORMIGA component
of BLUE STAR STING and by running the software SurfV, produce following table
(see
HELP on FORMIGA) (only amino acids with non zero ASA
are shown):
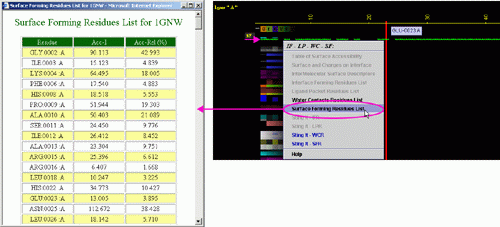
C) "STING it WCR
" will call a specific
Chime script and produce following 3D output - showing in cyan
CPK only amino acids with
an atom identified at the distance not larger than 4A from the co-crystalized
water: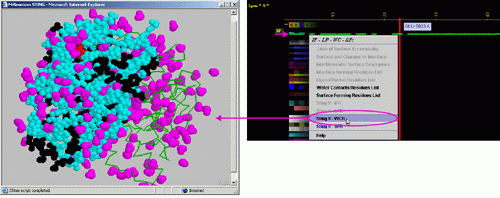
D) "STING it SFR
" will call a specific
Chime script and produce following 3D output - showing in red
CPK only SuRFace residues: