Electrostatic
Potential
[
ref. ]
Electrostatic Potential
is calculated using Delphi (12)
program according to the modifications done by Walter
Rocchia (13) and further adapted to JPD requirements (to be published).
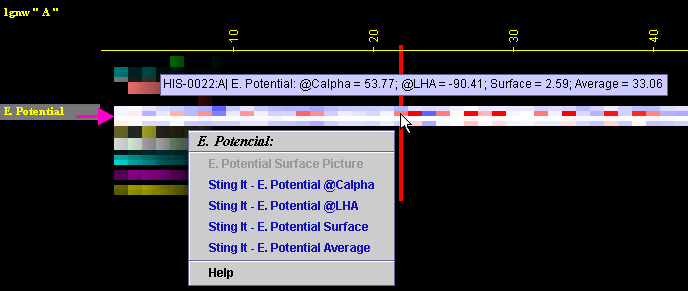
Placing the cursor above this element: pop-up area will show the position
(sequence number and AA three letter code) for selected amino acid and
the 4 numerical values for EP (@Calpha, @Last Heavy Atom (LHA),
@Surface patch closest to the selected residue and Average EP) for this
amino acid.
Left mouse click: no action
Right mouse
click: on any of the "EP" will generate following menu and
actions:
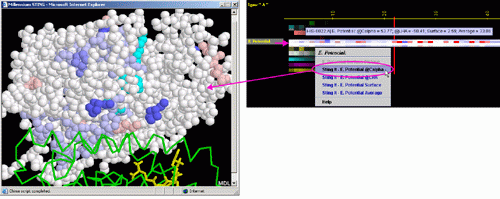
A) STING it "EP @Calpha": This option generates structural presentation
with color coding of the amino acids corresponding to the "EP"
color coding. Amino acids are presented in CPK rendering. EP at the
alpha carbon is presented.
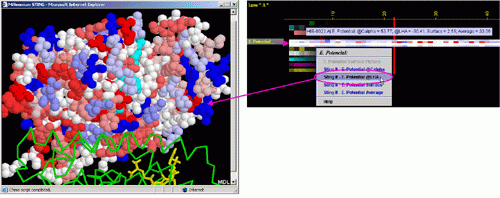
B) STING it "EP @LHA": This option generates structural presentation
with color coding of the amino acids corresponding to the "EP"
color coding. Amino acids are presented in CPK rendering. EP at the
Last Heavy Atom of the side chain is presented.
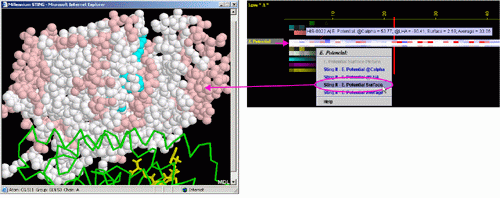
C) STING it "EP @Surface": This option generates structural
presentation with color coding of the amino acids corresponding to the
"EP" color coding. Amino acids are presented in CPK rendering.
EP at the Surface patch closest to the selected amino acid is presented.
D) STING it "EP Average": This option generates structural presentation
with color coding of the amino acids corresponding to the "EP"
color coding. Amino acids are presented in CPK rendering. The average
of EP values taken from all atoms of the selected amino acid is presented.
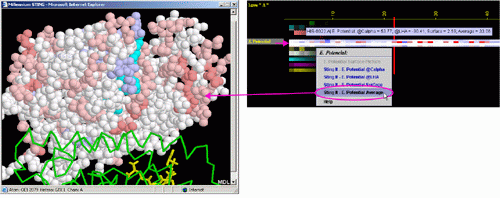
E) The EP values
shown at the picture below, were generated using GRASP
program. We use Delphi program from same lab (Barry
Hong's lab - Columbia University, NYC-USA). Figure below shows the
difference in concept when data display is in question: GRASP does it
one parameter at the time shown at the molecular surface. BLUE STAR STING
JPD does it all parameters at the same time for all amino acids (including
those at the surface). 