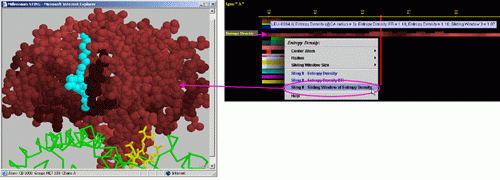
Entropy Density
The sum of values is calculated for the relative entropy (according to HSSP (7) data) of the amino acids encountered within the sphere of a given radius, and then divided by the volume of that sphere.
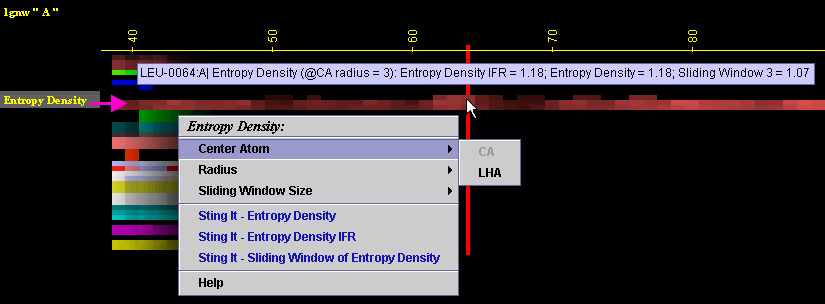
Placing the cursor above this element: pop-up area will show the position
(sequence number and amino acid three letter code) for selected amino
acid and the 3 numerical values for Entropy Density (@IFR, Internal and
Sliding window) for given amino acid.
Left mouse click: no action
Right mouse
click: on any of the "Entropy Density" will generate following
menu and actions:
A) A user may select either Ca or Last Heavy Atom (LHA) in the side chain
as a center of the sphere for calculating "Entropy Density"
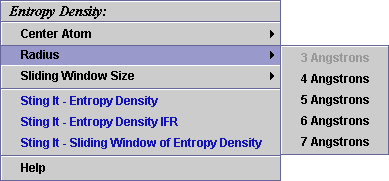
B) A user may select
among 5 possible radius values for the sphere:
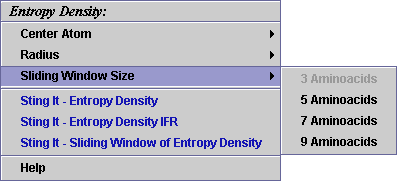
C) A user may select among 4 possible values for the sliding window size
(for the size of 5: a value for this parameter is collected over the central
residue and other 2 from the each side of the central one, summed and
divided by 5):
D) STING it "Entropy
Density": This option generates structural presentation with color
coding of the amino acids corresponding to the JPD row "Entropy Density"
color coding. The area of gaps (as well as ligands) is where the colors
are going to be different from the rest of the protein. Amino acids are
presented in CPK rendering.
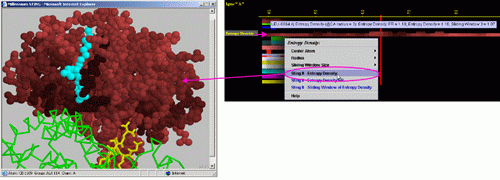
E) STING it "Entropy
Density IFR ": This option generates structural presentation with
color coding of the amino acids corresponding to the JPD row "Entropy
Density" color coding. The area of interface between the two chains
is where the colors are going to be different from the rest of the protein.
Amino acids are presented in CPK rendering.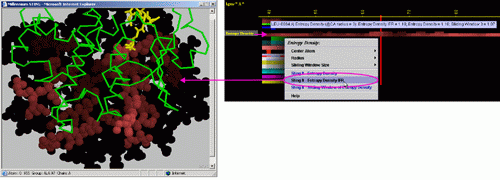
F) STING it "Sliding Window Entropy Density": This option generates
structural presentation with color coding of the amino acids corresponding
to the JPD row "Entropy Density" color coding. The area of gaps
is where the colors are going to be different from the rest of the protein.
Amino acids are presented in CPK rendering. A ligand here is presented
in cyan color.