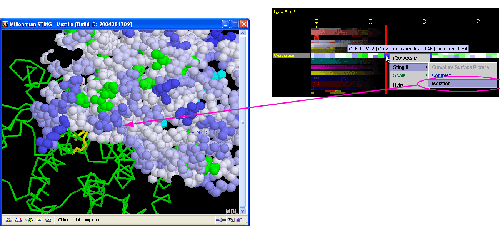
Curvature
The curvature values for each amino acid are calculated using the program SurfeRace (14). SurfeRace calculates the curvature value for each atom. The curvature of residues is the average of the surface atoms curvature.
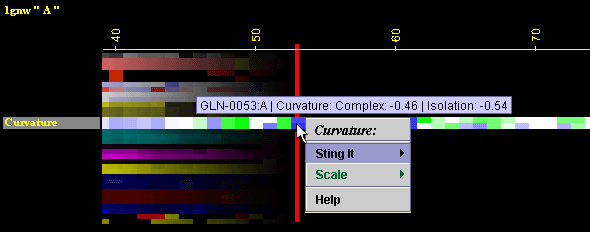
Placing
the cursor above this element: pop-up area will show the position
(sequence number and AA three letter code) for selected amino acid and
the numerical value for Curvature for this amino acid.
Left mouse click: no action
Right mouse
click: on any of the "Curvature" will generate following
menu and actions:
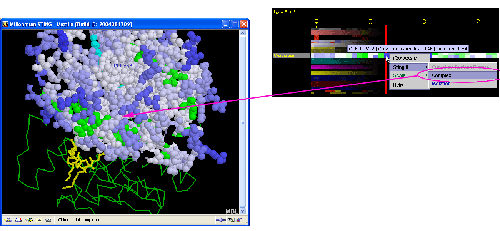
A) STING it : This option
generates structural presentation with color coding of the amino acids
corresponding to the "Curvature" color coding. Amino acids are
presented in CPK rendering. It is possible to show the Isolation or
Complex curvature
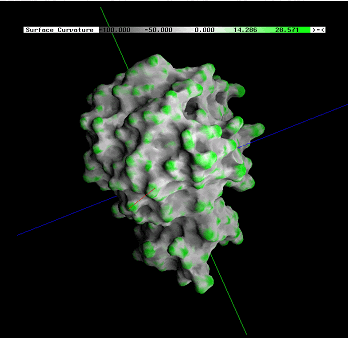
A protein
surface is not a plane surface. It does constitute of concave and convex
areas.
The concave portions of the surfaces, making a part of the pockets, are possible indicator of the surface regions considered probably for engaging in the interactions with the other proteins. In the docking process, two molecules should geometrically fit portions of the facing surfaces. The average curvature of that surface portion should be equal at the two interfaces but opposite in sign.
![]()

| Figure at the right side shows the difference in concept when data display is in question: GRASP does it one parameter at the time shown at the molecular surface. BLUE STAR STING JPD does it all parameters at the same time for all amino acids (including those at the surface). |
 |