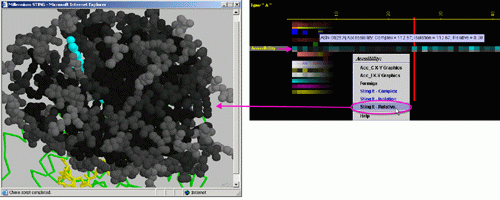
Accessibility
The
Amino acid accessibility is calculated according to SurfV
(10) program. JPD shows 3 values: for the protein chain in isolation,
for the protein chain in complex with the other chain (if) present in
the PDB file and finally, a relative accessibility (the last one given
by the table of absolute solvent
accessible area for amino acids).
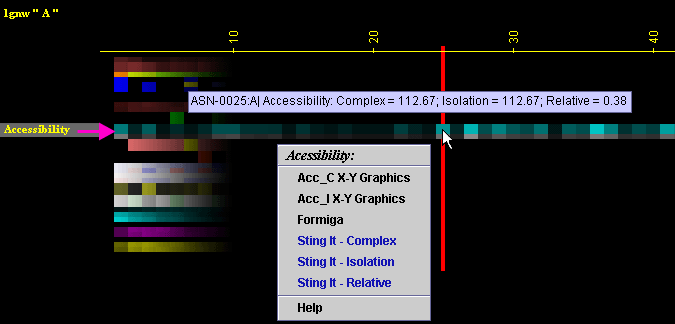
Placing the cursor above this element: pop-up area will show the position
(sequence number and AA three letter code) for selected amino acid and
the Accessibility values for the chain in complex with the other chain,
for the chain in isolation and for the relative accessibility.
Left mouse click: no action
Right mouse
click: on any of the "Accessibility" will generate following
menu and actions:
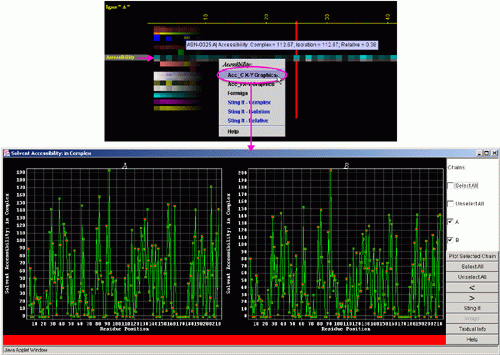
A) "Accessibility X-Y Graphics" will produce following Java
X-Y graphics (X= sequence number, Y= Accessibility in Complex) with the
interactive possibilities described in dedicated
HELP manual:
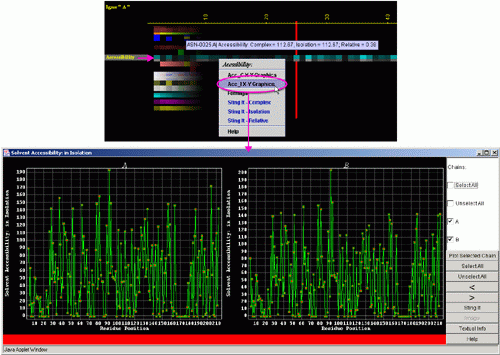
B) "Accessibility X-Y Graphics" will produce following Java
X-Y graphics (X= sequence number, Y= Accessibility in Isolation) with
the interactive possibilities described in dedicated
HELP manual:
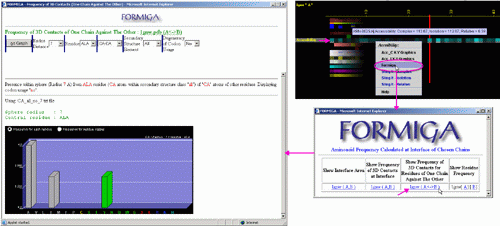
C) This option will call FORMIGA component of BLUE STAR STING and by running
the software SurfV, produce following output (see
HELP on FORMIGA):
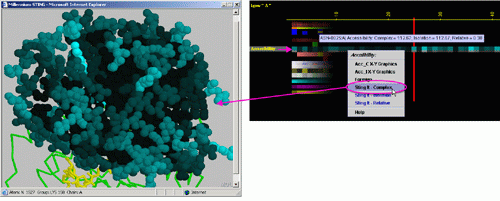
D) "STINGit - Complex" will induce CHIME window to show the
values of accessibility in color coding corresponding to the same color
code used by this specific JPD row. The accessibility displayed is for
the protein chain in complex with the other chain (if) present in the
PDB file.
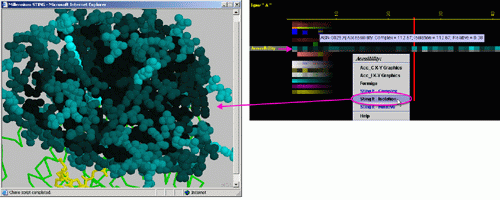
E) "STINGit - Isolation" will induce CHIME window to show the
values of accessibility in color coding corresponding to the same color
code used by this specific JPD row. The accessibility displayed is for
the protein chain in isolation.
F) "STINGit - Relative" will induce CHIME window to show the
values of Relative accessibility in color coding corresponding to the
same color code used by this specific JPD row. The accessibility displayed
is for the relative accessibility calculated as a ratio of the value for
the accessibility of a selected amino acid belonging to the protein chain
in isolation and divided by the value given in the table
of absolute solvent accessible area for amino acids which basically
shows SurfV output with the calculated accessibility of a single amino
acid standing in the solvent all by itself.